Case01 Boundary Optimisation
In order to perform the inverse analysis a folder structure with a template datafile (that will be used to generate all the models data files), the data used as target results need to be defined in addition to the main data file (.inp) defining the inverse analysis data. The starting folder structure with the data required is provided in MEM_003 \Case01\Data and is as follows:
•Target: folder containing the target results file(s) to be used for optimisation of the boundary condition.
▪MEM_003_Ref_001.hdh
▪MEM_003_Ref_002.hdh
▪MEM_003_Ref_003.hdh
•Template: folder containing the template data file(s) that will be used to define all models simulation data files.
▪MEM_003_Case01.contact
▪MEM_003_Case01.dat
▪MEM_003_Case01.geo
▪MEM_003_Case01.geometry
▪MEM_003_Case01.mat
▪MEM_003_Case01_Porosity.spat
▪MEM_003_Case01_PP_Initial.spat
•Test: "Empty" folder to which the simulation results will be written to.
•MEM_003_Case01.inp: Data defining the inverse analysis.
For the present tutorial example the user is assumed to be familiar with ParaGeo and only the data relevant to the inverse analysis and boundary optimisation will be discussed in detail.
MEM_003_Case01.inp file
The MEM_003_Case01.inp file is the main file in which the data associated to the inverse modelling / optimisation procedure is defined. The data structures and all their corresponding keywords defined within this file are described in the GeoInv Data Structures manual section.
Application_data
The Application_data data structure is used to define main controls associated to the code and algorithm used for the inverse modelling procedure.
Boundary_opt_parameters
The Boundary_opt_parameters is the data structure used to define the type of boundary condition to be optimised, the initial value for the boundary condition and the allowed range of values for the boundary condition in the models to be generated by the optimisation procedure. This data is going to be used to populate the blank Parameterised_boundary data structure defined within the template data file.
NA_options
The NA_options data structure is used to set the options for the Nearest Neighbour Algorithm.
File_data
File_data data structure is used to define the name of the template data file as well as the directories for the template, target solution and test folders.
Misfit_data_set
The Misfit_data_set data structure is used to define the files and variables to be used as a target for the optimisation procedure. Note that a Misfit_data_set data structure must be defined for each target solution file to be used for the optimisation procedure.
Inverse_case
The Inverse_case data structure is used to identify the Misfit_data_set data structures (by number) defined in the present .inp file that will be active for the present optimisation procedure.
|
Template: Data File Description
The template folder should contain the template data that will be used to generate all models. The template data basically consists of the full data to define a single simulation except the Parameterised_boundary data which is left in blank and will be populated by ParaGeoInv with different boundary displacement values for every model generated during the optimisation procedure. Another difference is that in the template it is generally recommended to set Control_data with no plot file output in order to save CPU time, so that plot files may be obtained from an additional run of the optimised model.
The present example involves two simulation stages which consist of gravity initialization and tectonic displacement respectively. The basic data comprises:
1.Geometry and mesh data defined within the .geo file. 2.Geometry_set data for all model boundaries, stratigraphy horizons and fault surfaces. 3.Group_data and Group_control_data for the seven formations. 4.Contact_global, Contact_set, Contact_property, Contact_surface and Fault_set defining data for the fault. It should be noted that the contact remains elastic during the initialisation stages considered in the present case. 5.Material_data within the .mat file defining properties for all formations. 6.Stratigraphy data to identify the top surface and define its conditions (Stratigraphy_definition, Stratigraphy_horizon and Stratigraphy_surface_load). 7.Support_data defining displacement constraints normal to the model boundaries. 8.Gravity_data with the corresponding Time_curve_data. 9.Initialisation data: a.Geostatic_data and a Spatial_grid to initialise porosity values b.Spatial_grid and Spatial_boundary to initialise pore pressure values c.Data for initialisation of temperature by applying a temperature gradient vs depth (Global_loads, Spatial_variation_definition, Spatial_variation_values, Load_case_control_data) d.Geostatic_control_data assigning the appropriate initialisation conditions at every stage defined within the geostatic.set file. 10. Control_data for an implicit simulation.
History points
Parameterised Boundary
Control_data
|
Results
The results for the present case are provided in MEM_003\Case01\Results.
The present inverse analysis has necessitated running 30 models to achieve convergence (initial sample of 6 models + four resamples of 6 models). Five main result files have been output from the inverse analysis:
•MEM_003_Case01.log: File with log of the operations performed during the inverse analysis.
•MEM_003_Case01.ParAll: File containing the model number, parameter (boundary displacement) value and misfit value for all models.
•MEM_003_Case01.ParBest: File containing the model number, parameter (boundary displacement) value and misfit value for the best 5 models (as specified in Number_models_output keyword within NA_options data structure).
•MEM_003_Case01.ParOpt: File containing the model number, parameter (boundary displacement) value and misfit value for the best / optimal model.
•MEM_003_Case01.out: File with comparison of the target and model history results for the optimal model and the best 5 models (as specified in Number_models_output keyword within NA_options data structure). The last set of output at the end of the file contain the model number, parameter (boundary displacement) value and misfit value for the 30 models run during the inverse analysis. The format of the file is illustrated below with some comments inserted here for clarification of the content in each section of the data file.
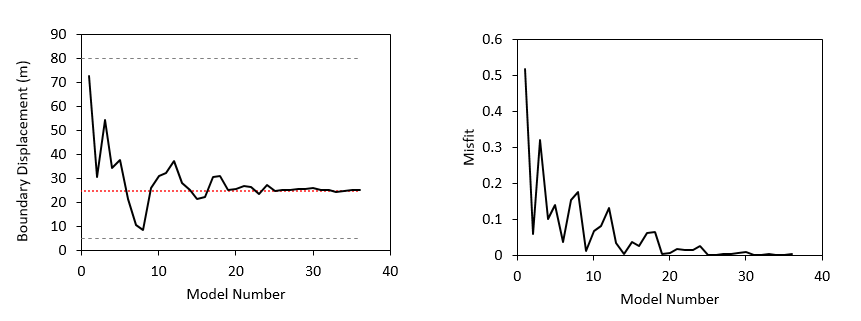
In the present inverse analysis the optimal model corresponds to model number 26 with an optimised boundary displacement of 25.03 m and a misfit value of 0.3624·10-3. Note that in the reference solution the imposed displacement was -25.0 m which means that the present inverse analysis has found an optimal displacement which only has 0.12 % of error relative to the one prescribed in the reference solution. In the following figure it is shown the evolution of the tested boundary displacements and the corresponding misfit values during the inversion analysis.
Tested boundary displacements for each model (left) and corresponding misfit value (right). In the figure in the left the value used in the reference solution is shown with the dotted red line whereas the imposed bounds for the inverse analysis are shown in discontinuous grey lines.
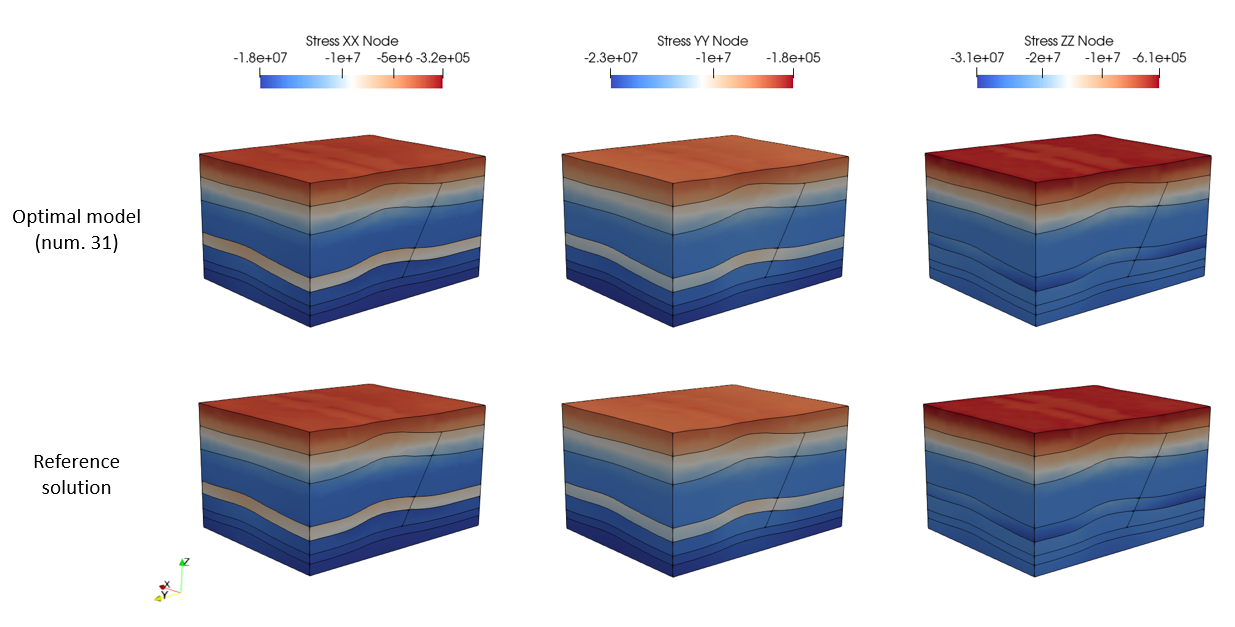
In the folder MEM_003\Case01\Results\Test the data files for all the models run during the inversion analysis are preserved. The optimal model (number 26) has been manually copied and run into the folder MEM_003\Case01\Results\Optimal_Run. The figure below shows the comparison of the optimal model results and the results from the reference solution. As expected the results are almost identical.
Stresses plot file comparisons between the optimal model and the reference solution
|